Table S1 for Spirodela polyrhiza Species
The table shows the EXPs of Spirodela polyrhiza species. You can filter by EXP Id.
| Order | Species Name | EXP Id | Genom DB Id | Chromosome | Start Position | End Position | Protein Length | PI | Molecular Weight | Instability Index | Stable or Unstable | Ncbi Id | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Spirodela polyrhiza | SpEXPA-01 | Spipo1G0052100 | pseudo1 | 3369307 | 3370575 | 241 | 7.62 | 26317.01 | 33.12 | Stable | None | |||||
| 2 | Spirodela polyrhiza | SpEXPA-02 | Spipo12G0039500 | pseudo12 | 2914176 | 2915132 | 255 | 7.08 | 27159.43 | 39.01 | Stable | None | |||||
| 3 | Spirodela polyrhiza | SpEXPA-03 | Spipo17G0044900 | pseudo17 | 3250152 | 3251155 | 279 | 9.69 | 30602.51 | 42.72 | Unstable | None | |||||
| 4 | Spirodela polyrhiza | SpEXPA-04 | Spipo20G0037100 | pseudo20 | 3105648 | 3106573 | 253 | 8.92 | 26426.98 | 31.3 | Stable | None | |||||
| 5 | Spirodela polyrhiza | SpEXPA-05 | Spipo22G0026300 | pseudo22 | 1967764 | 1968637 | 251 | 8.26 | 26726.03 | 33.8 | Stable | None | |||||
| 6 | Spirodela polyrhiza | SpEXPA-06 | Spipo22G0037800 | pseudo22 | 2555503 | 2556869 | 260 | 9.38 | 27891.84 | 29.85 | Stable | None | |||||
| 7 | Spirodela polyrhiza | SpEXPA-07 | Spipo23G0016200 | pseudo23 | 960680 | 961626 | 243 | 9.2 | 26028.42 | 38.62 | Stable | None | |||||
| 8 | Spirodela polyrhiza | SpEXPB-01 | Spipo1G0037900 | pseudo1 | 2547012 | 2548753 | 262 | 6.3 | 28131.15 | 36.74 | Stable | None | |||||
| 9 | Spirodela polyrhiza | SpEXPB-02 | Spipo11G0034600 | pseudo11 | 2839496 | 2840837 | 277 | 6.18 | 29338.15 | 56.67 | Unstable | None | |||||
| 10 | Spirodela polyrhiza | SpEXPB-03 | Spipo11G0034700 | pseudo11 | 2848658 | 2849894 | 265 | 8.95 | 27358.96 | 37.17 | Stable | None | |||||
| 11 | Spirodela polyrhiza | SpEXLA-01 | Spipo1G0075200 | pseudo1 | 4671213 | 4672358 | 265 | 8.75 | 28638.68 | 38.97 | Stable | None | |||||
| 12 | Spirodela polyrhiza | SvEXPA-35 | Sevir.9G540000.1.p | Chr9 | 5.346114E+07 | 5.346248E+07 | 250 | 8.88 | 26928.55 | 26.92 | Stable | XP_034573009.1 | |||||
| Table S1 Count: 12 | |||||||||||||||||
Click on the pictures to see
| Gene Structure | Chromosomal Location | Conserved Motif | ||
|---|---|---|---|---|
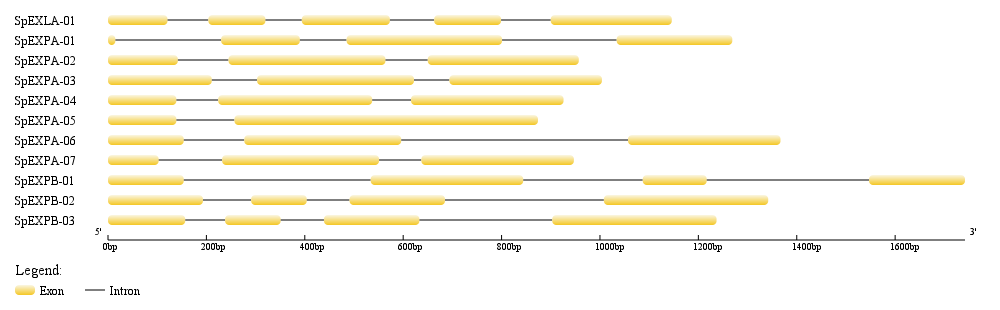 |
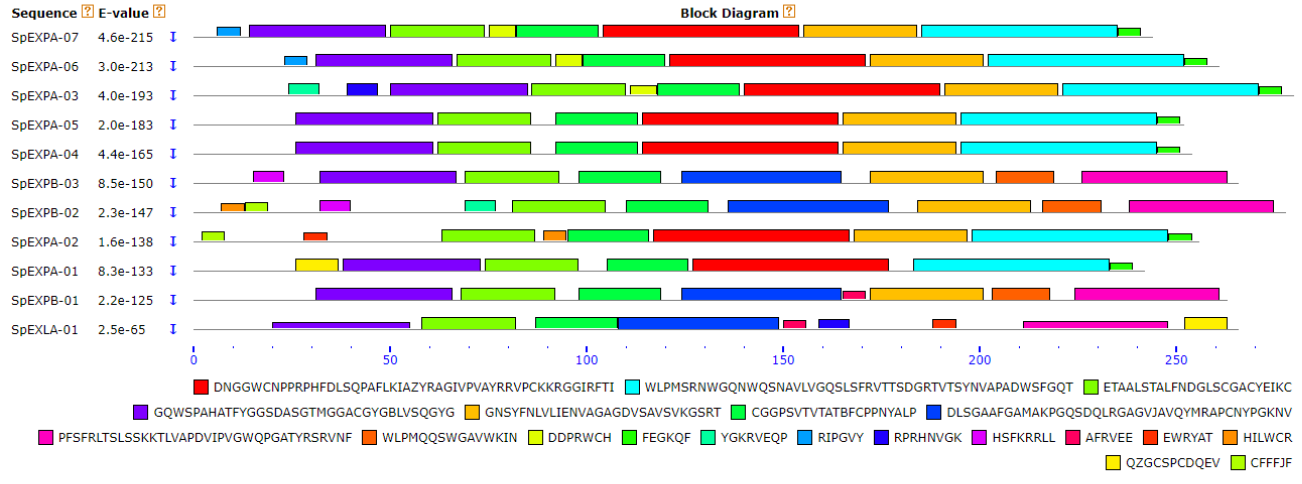 |
|||
| Image Count: 1 | ||||
| Article Link | ||||
|---|---|---|---|---|
| https://www.nature.com/articles/ncomms4311 | ||||
| Link Count: 1 | ||||
×
![]()